Reconstructing a Phylogenetic Tree Using SNP Information from WGS Data
Phylogenetic analysis is a key part of evolutionary biology, allowing researchers to infer the relationships between species, populations, or strains.
By using whole genome sequencing (WGS) data, we can identify single nucleotide polymorphisms (SNPs) and use them to construct high resolution phylogenetic trees.
In this blog post, I’ll guide you through reconstructing the phylogenetic tree of Rhynchosporium commune, a significant fungal pathogen of barley crops, using SNP information obtained from WGS data. This tutorial provides a practical, step-by-step guide from raw data processing through to tree visualisation.
Environment Setup
Before we start, we’ll need to set up the environment and install all of the required software.
The scripts used in this tutorial can be found in my GitHub repository, and are written to be run on a Slurm-based HPC cluster. If you have Conda installed, you can install all of the software in a new environment as below:
# Clone repository
git clone https://github.com/rj-price/rhynchosporium_tree.git
cd rhynchosporium_tree
# Create conda environment
conda create -n snp_tree \
ncbi-datasets-cli \
fastqc \
multiqc \
trimmomatic \
python \
pandas \
biopython \
bwa \
samtools \
bcftools \
raxml-ng
# Activate conda environment
conda activate snp_tree
Data Preparation
Download WGS Data and Reference Genome
With the environment set up, the next step involves acquiring the raw sequencing data and a reference genome.
For this analysis, I searched the Sequence Read Archive (SRA) for Rhynchosporium WGS data and downloaded the available metadata (SraRunTable.txt).
I then used pre-generated scripts via the European Nucleotide Archive (ENA) to downloaded all of the Rhynchosporium reads from the BioProject accessions PRJNA327656 and PRJNA419548:
# Create directory for raw reads
mkdir reads && cd reads
# Download WGS data using pre-generated scripts
bash ../scripts/ena-file-download-selected-files-20240216-1125.sh
bash ../scripts/ena-file-download-selected-files-20240216-1138.sh
For the reference genome, the NCBI Datasets tool was used to download the R. commune UK7 genome:
# Create directory for reference genome
mkdir UK7 && cd UK7
# Download UK7 genome
datasets download genome accession GCA_900074885.1 --filename GCA_900074885.1.zip
# Unzip, move and rename
unzip GCA_900074885.1.zip
mv \
ncbi_dataset/data/GCA_900074885.1/GCA_900074885.1_version_1_genomic.fna \
R_commune_UK7.fasta
Quality Control of Sequencing Data
Downstream analyses rely on high quality sequencing data. To assess the quality of the raw reads, I used FastQC, followed by MultiQC to summarise the results:
# Run FastQC on raw data
cd reads
for file in *gz; do
sbatch ../scripts/fastqc.sh $file
done
# Aggregate results with MultiQC
multiqc fastqc/
Adapters and low quality sequences were trimmed using Trimmomatic based on the QC report, followed by a second round of quality control:
# Create directory for trimmed reads
mkdir trimmed && cd trimmed
# Trim reads using Trimmomatic
for fwd in ../reads/*_1.fastq.gz; do
rev=$(ls $fwd | sed 's/_1/_2/g')
sbatch ../scripts/trimmomatic_pe.sh $fwd $rev
done
# QC on trimmed reads
for file in *gz; do
sbatch ../scripts/fastqc.sh $file
done
multiqc fastqc/
Finally, to facilitate easy comparisons across samples, a custom python script was used to rename the reads using a custom naming convention based on the available SRA metadata:
mkdir renamed_reads
python scripts/rename_reads.py SraRunTable.txt trimmed/ renamed_reads/
Phylogenetic Tree Reconstruction
Mapping Reads to the Reference Genome
To identify SNPs, the reads were aligned to the UK7 reference genome using BWA. After indexing the genome, the reads were mapped, and the alignments were sorted and indexed using Samtools:
# Index the genome
bwa index UK7/R_commune_UK7.fasta
# Map reads to the genome
mkdir alignment && cd alignment
for fwd in ../renamed_reads/*_F.fastq.gz; do
rev=$(ls $fwd | sed 's/_F/_R/g')
sbatch ../scripts/bwa-mem.sh ../UK7/R_commune_UK7.fasta $fwd $rev
done
# Sort and index alignments
for file in *sam; do
sbatch ../scripts/samtools_sort.sh $file
done
Variant Calling and Filtering
After generating a pileup of aligned reads, variants were called using bcftools. These were then filtered for high-quality SNPs based on quality scores, retaining only the highest-confidence SNPs for phylogenetic analysis:
# Index genome
samtools faidx UK7/R_commune_UK7.fasta
# Generate pileup and call variants
mkdir variant_calling && cd variant_calling
for file in ../alignment/*bam; do
realpath $file >> rhynchosporium.list
done
sbatch ../scripts/bcftools_call.sh ../UK7/R_commune_UK7.fasta rhynchosporium.list
# Filter variants
bcftools view \
rhynchosporium.vcf -e 'QUAL<800' --types snps \
> rhynchosporium_highqual.vcf
Preparing Data for Phylogenetic Analysis
The filtered SNP data in VCF format was converted to PHYLIP format using the vcf2phylip.py script from https://github.com/edgardomortiz/vcf2phylip and invariant sites were removed using the ascbias.py script from https://github.com/btmartin721/raxml_ascbias:
# Convert VCF to PHYLIP
python ../scripts/vcf2phylip.py -i ./rhynchosporium_highqual.vcf
# Remove invariant sites
python ../scripts/ascbias.py -p ./rhynchosporium_highqual.min4.phy -o rhynchosporium_highqual.phy
Building the Phylogenetic Tree
With the prepared dataset, I used RAxML-ng for phylogenetic inference:
sbatch ../scripts/raxml.sh rhynchosporium_highqual.phy
Visualising the Tree
The final phylogenetic tree was visualised using iTOL, a useful online tool for exploring and annotating trees. Below are examples of the visualisations generated:
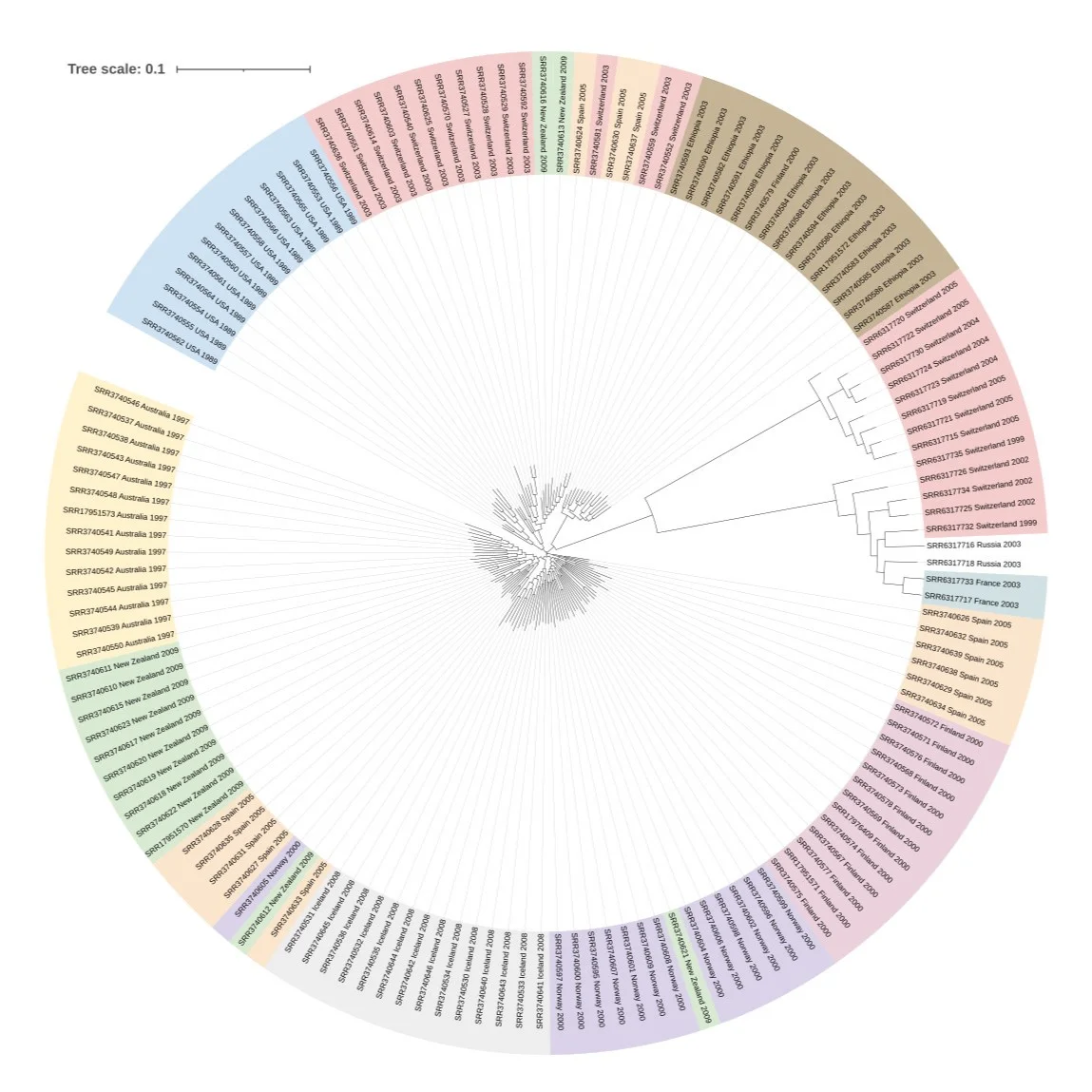
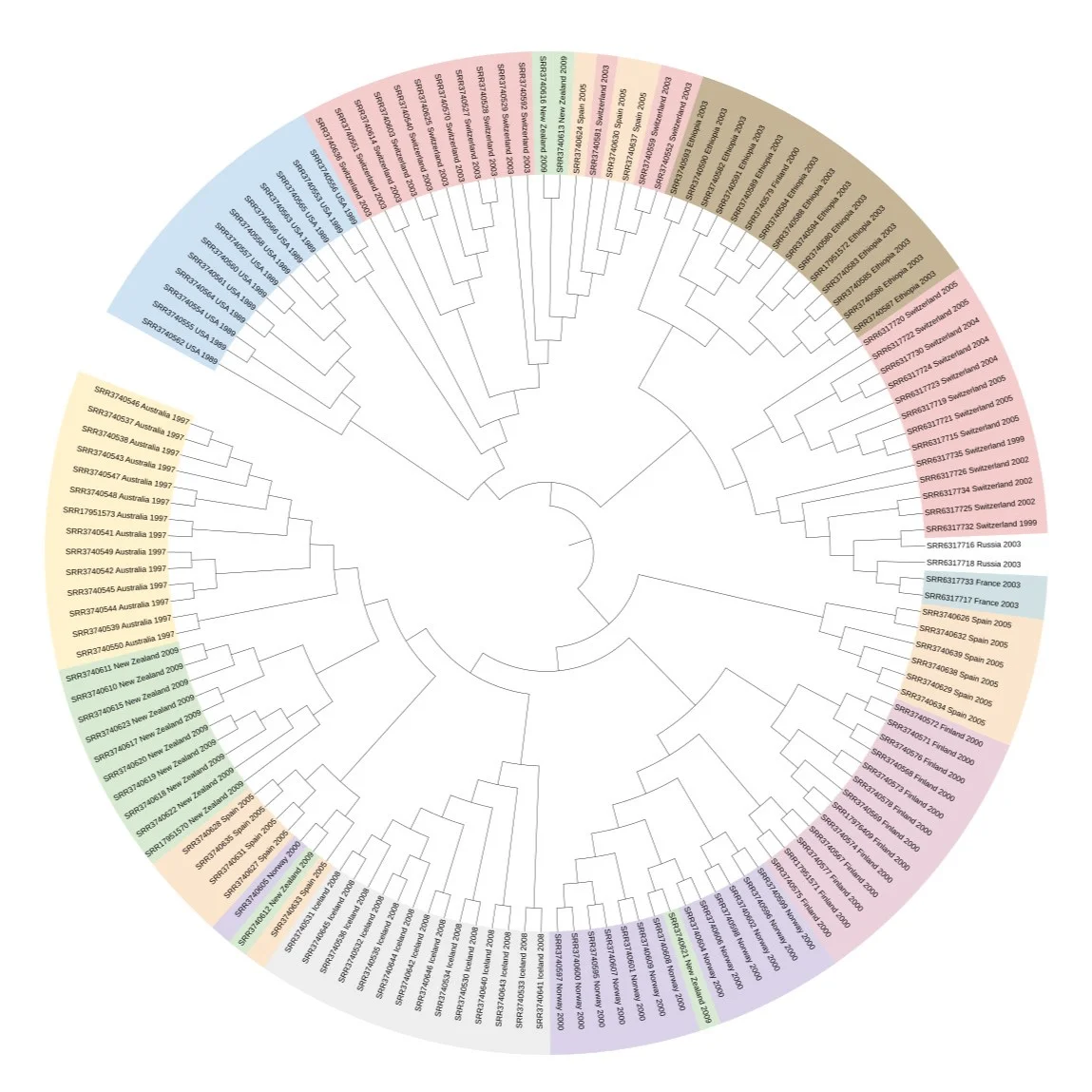
Summary
Reconstructing a phylogenetic tree from WGS data is a multi-step process involving data preparation, SNP calling, and tree inference. This guide outlines the workflow for analysing Rhynchosporium commune WGS data but can be adapted for other species or datasets.
By following these steps, you can uncover evolutionary insights and explore the genetic diversity of your own organisms of interest.
Support
If you find this blog post helpful, please consider buying me a coffee on Ko-fi. Your support is very much appreciated!
Enjoy Reading This Article?
Here are some more articles you might like to read next: