Fusarium Plant Pathogens - Comparative Genomics
Uncovering the molecular toolkit of lettuce wilt pathogens.
Fusarium oxysporum f. sp. lactucae (Fola) is the causal agent of lettuce Fusarium wilt, a devastating disease that poses a significant threat to production worldwide. While Race 1 (Fola1) has long been a problem in warmer climates, the recent emergence and spread of the highly aggressive Race 4 (Fola4) across Northern Europe has created a new and urgent challenge for the industry. Our work uses comparative genomics and transcriptomics to dissect the genetic differences between these economically important races, revealing how they have evolved and identifying the molecular tools they use to cause disease.
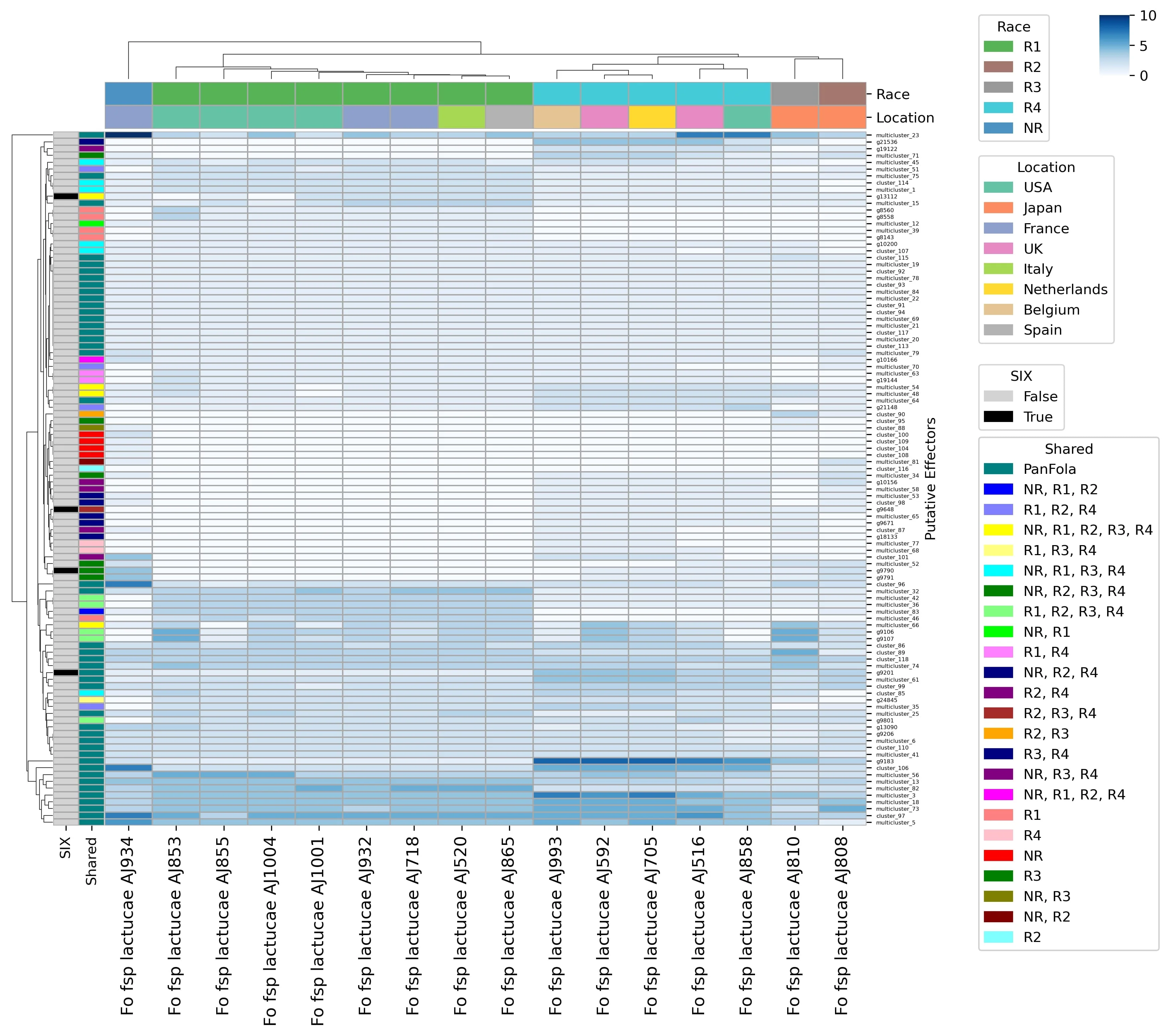
To understand what makes these races distinct, we conducted the first ever genome and transcriptome analyses for Fola1 and Fola4. Our phylogenomic analysis revealed that Fola4 did not simply evolve from the older Fola1. Instead, they are sister clades, meaning they diverged from a common ancestor and followed separate evolutionary paths. This was further supported by the lack of large-scale structural similarity (synteny) between their accessory genomes, the highly variable parts of the genome that carry genes for virulence.
This research also uncovered evidence for two distinct variants existing within Race 4, which differ in their effector profiles, SIX gene sequences, and genome structure. The data strongly suggest these variants acquired their key virulence genes independently, likely through the horizontal transfer of chromosomes from different F. oxysporum donors.

The ability of a pathogen to infect its host is determined by a suite of proteins called effectors. Our analysis showed that the effector repertoires of Fola1 and Fola4 are significantly different, with several effector genes being unique to each race.
The differences were particularly clear within a critical family of effectors known as Secreted In Xylem (SIX) genes:
- SIX8: This gene is completely absent in Fola1 but is present and highly expressed by Fola4 during lettuce infection. Its presence is a key genetic marker for Race 4.
- SIX14: While present in the genomes of both races, the copies in Fola1 have been disrupted and silenced by the insertion of mobile genetic elements (transposons). In contrast, the single copy in Fola4 is undamaged and highly expressed during infection, making its activity another key differentiator between the races.
These discoveries pinpoint specific genes that likely determine the different virulence profiles of Fola1 and Fola4, providing clear targets for molecular diagnostics and informing the development of new resistant lettuce varieties. Building on this foundation, our ongoing research is expanding this comparative genomics approach to include all four known Fola races, as well as unique field variants. This broader analysis aims to map the pathogen’s full diversity, helping us to better predict its evolution and develop more durable disease management strategies.