Raspberry Breeding - Plant Genomics
Enhancing raspberry breeding through cultivar genomics.
Red raspberry (Rubus idaeus) is an economically important soft fruit crop, but breeding new varieties with improved traits is a slow and complex process. This is largely due to the species’ highly heterozygous genome, which has historically made genetic analysis challenging.
This research programme uses genomics to overcome these hurdles, creating foundational resources for the community and using them to dissect the genetic basis of key agricultural traits.
Building the Genomic Toolkit
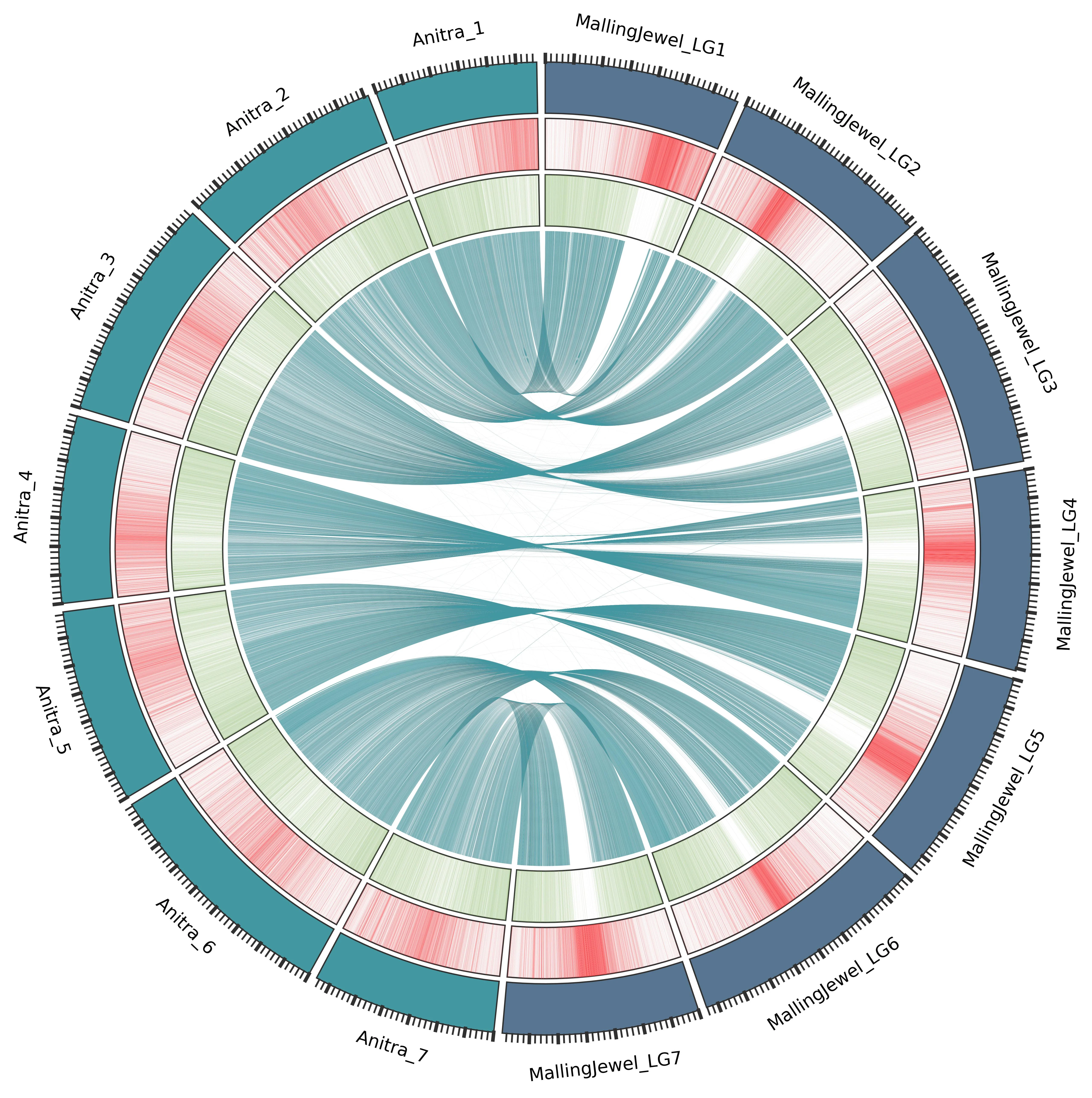
A central part of our work involves constructing high-quality, chromosome-scale genome assemblies for key raspberry cultivars, such as the primocane ‘Autumn Bliss’ and the floricane ‘Malling Jewel’.
By using long-read sequencing technologies like Oxford Nanopore, we can generate highly contiguous assemblies that are more complete than those produced with older methods alone.
This approach is particularly effective at resolving complex, highly repetitive DNA regions, such as the centromeres, which were not well-defined in previous genome sequences. These vastly improved and well-annotated genomes, serve as an essential toolkit for both fundamental research and applied breeding efforts.
Uncovering the Genes Behind Key Traits
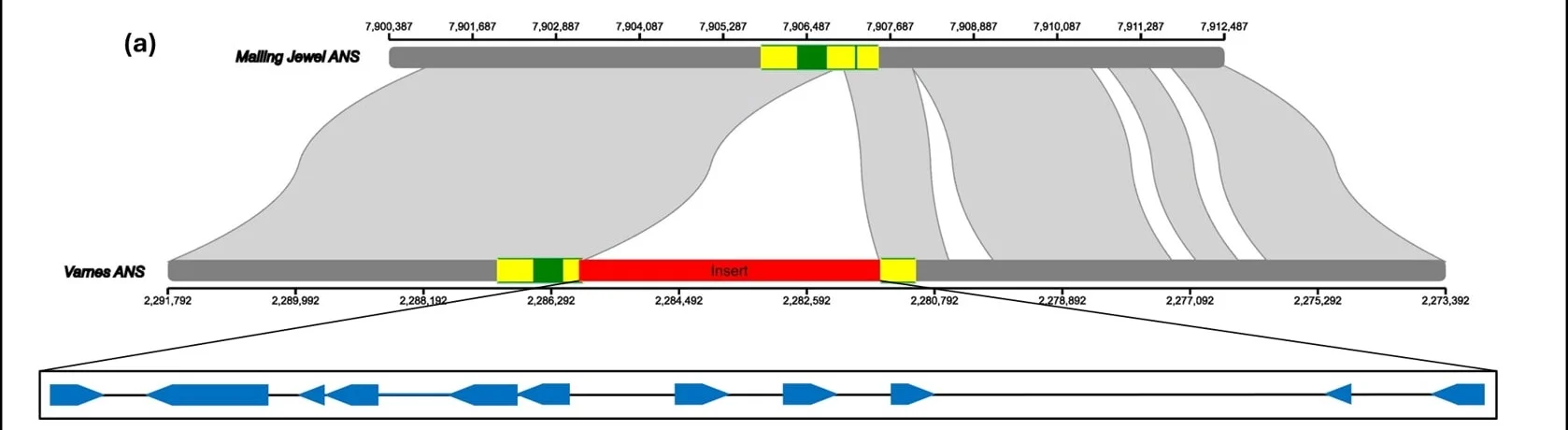
With these high-quality genomic resources in hand, we can effectively investigate the molecular basis of important traits. In one project, we used a multi-omics approach to understand the cause of the apricot fruit colour in the raspberry cultivar ‘Varnes’.
Metabolomic analysis confirmed that apricot-coloured fruits produce significantly lower levels of anthocyanin pigments compared to red varieties. By sequencing the ‘Varnes’ genome, we identified the genetic cause, a 4.3 kb CACTA-like transposable element had inserted itself into the Anthocyanidin synthase (Ans) gene, a key enzyme in the pigment production pathway. This insertion creates a truncated, dysfunctional protein, explaining the reduced fruit colour.
We confirmed that ‘Varnes’ carries two copies of this mutated gene, while also discovering that an unrelated, red-fruited cultivar, ‘Veten’, carries one copy, acting as a silent carrier for this recessive trait.
This work not only solves a long-standing question in raspberry genetics but also provides breeders with a precise molecular marker to select for fruit colour in their programmes. This demonstrates how foundational genomics work directly enables the discovery of genes controlling valuable agronomic traits.